New Version Update:
A version 2 of OTUX (OTUXv2) has been released. This approach utilizes the ‘amplicon specific’ sequences, when compared to ‘V-region specific’ sequences in OTUX. Please check the OTUXv2 page for more details.
The most common approach for analysis in 16S data is to perform taxonomic assignment using the RDP classifier. However, when we perform taxonomic assignment, some part of the sequence is unclassified. These ‘unclassified sequences’ may even scale up to 70% of the total, and hence is not included for any further analysis. To overcome this issue researchers refine to clustering the sequences into ‘Operational Taxonomic Units’ or OTUs. Conventional OTU picking approaches suffer from certain limitations. While DNA sequencing errors can lead to an increased number of detected OTUs, the use of different clustering approaches may result in formation of alternate OTU clusters. Also, given the limitations pertaining to read-lengths generated by current generation of short-read sequencing techniques, the targeted amplicon often consists of a selected region from the phylogenetic marker gene, instead of the entire gene. Since the reference databases catalogue full length marker genes, querying the same with ‘short’ sequence reads for OTU identification/ taxonomic classification can yield suboptimal results. It may also be noted that rate of evolution (accumulation of mutations) is not always uniform across the length of a chosen marker gene (or in its variable regions) across different taxonomic clades. This may lead to significantly different OTU clustering results based on the choice of the target region. In summary, while OTUs identified/ classified using reference based methods vs de novo clustering methods can provide different results, any comparison between the results obtained from studies utilizing different variable regions of a given marker gene also loses relevance.
The OTUX database and associated OTU picking approach intends to overcome the above limitations by using customized reference OTU databases which are specific to the targeted regions of the chosen marker gene (such as 16S rRNA). The OTUX (meta)database consists of 19 distinct OTU databases corresponding to the different stretches of variable regions (V-regions) of the bacterial 16S rRNA gene, that are commonly targeted for amplicon sequencing in microbiome studies. Each of the V-region specific databases consists of OTUs (OTUX-OTUs) identified by clustering sequence fragments from corresponding stretches of V-regions cropped out from full-length 16S rRNA gene sequences catalogued in reference databases. In addition, a ‘mapping matrix’ is also presented which lists the probabilities of association of any of the OTUX-OTUs to the reference OTUs present in the widely used Greengenes OTU database (consisting full-length marker genes). An open-reference based OTU picking approach against an appropriately selected OTUX V-region database allows obtaining results similar to de novo OTU clustering results. Further, using the mapping matrix, the OTU abundance profiles obtained in terms of OTUX-OTUs can be ‘mapped back’ and represented in terms of Greengenes OTUs. Mapping back enables comparing OTU-picking/ taxonomic annotation results from different microbiome studies, even if the choice of targeted V-regions had been different. The utility of the OTU picking approach using OTUX database has been extensively validated with multiple simulated sequence datasets mimicking microbiome samples collected from diverse environments.
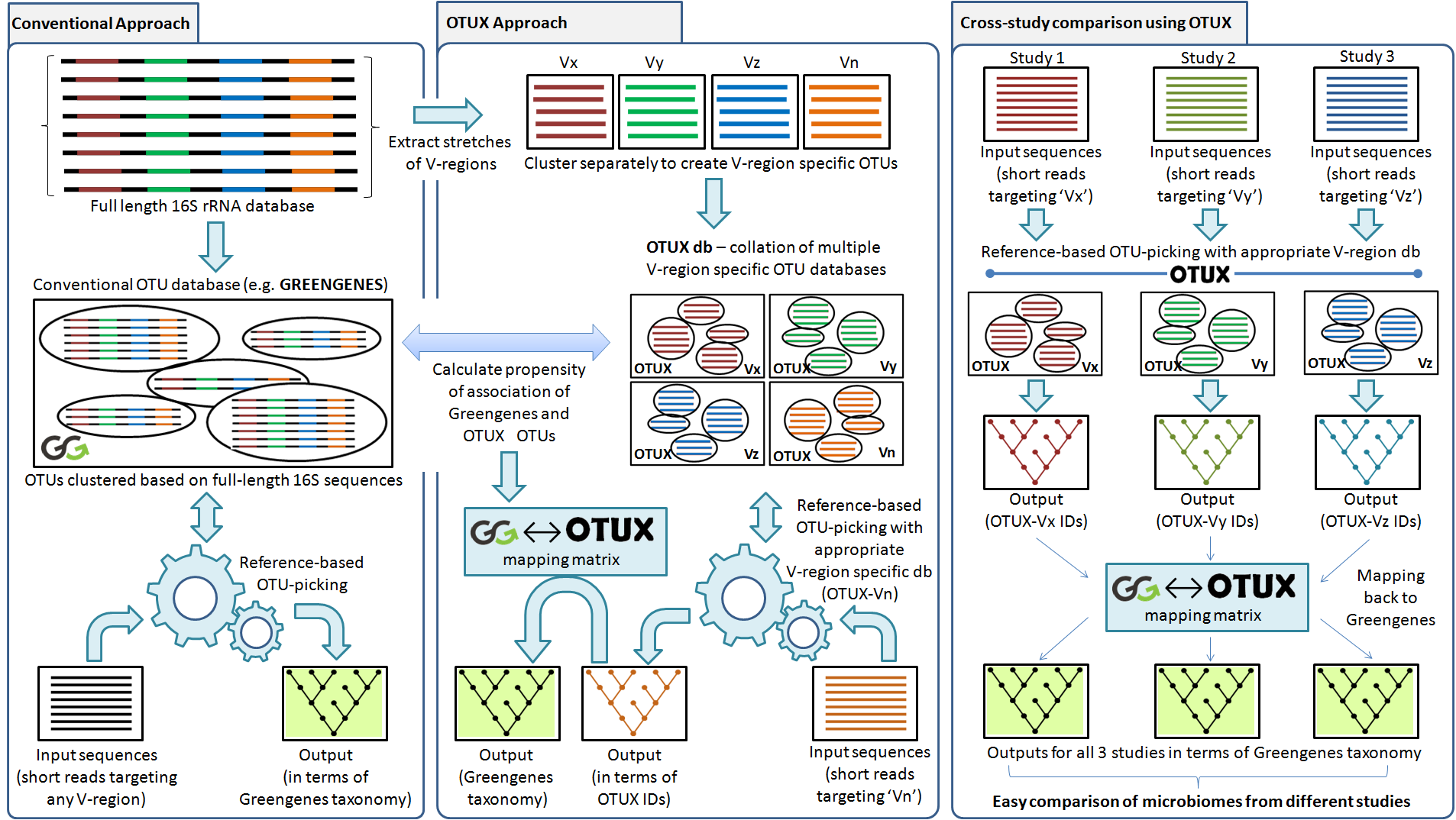
OTUX approach:
Deepak Yadav, Anirban Dutta, Sharmila S Mande, OTUX: V-region specific OTU database for improved 16S rRNA OTU picking and efficient cross-study taxonomic comparison of microbiomes, DNA Research, Volume 26, Issue 2, April 2019, Pages 147–156, https://doi.org/10.1093/dnares/dsy045
OTUXv2 approach:
Manuscript submitted
"OTUX" SOFTWARE TOOL IS NOT INTENDED TO BE USED FOR TREATING OR DIAGNOSING HUMAN SUBJECTS.
"OTUX" or any documents available from this server ARE PROVIDED AS IS WITHOUT ANY WARRANTY OF ANY KIND, EITHER EXPRESS, IMPLIED, OR STATUTORY, INCLUDING, BUT NOT LIMITED TO, ANY IMPLIED WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND FREEDOM FROM INFRINGEMENT, OR THAT "OTUX" or any documents available from this server WILL BE ERROR FREE.
In no event will the authors, their employers or any of the lab/office members be liable for any damages, including but not limited to direct, indirect, special or consequential damages, arising out of, resulting from, or in any way connected with the use of "OTUX" or documents available from this server.
The authors will try their best to maintain the privacy and confidentiality of the uploaded user data and will not use the data for any work directly or indirectly except for software debugging purpose.
