The OTUX version 2 (OTUXv2 ) approach utilizes an "amplicon specific" reference OTU database (whereas the OTUX approach utilizes the "V-region specific" reference OTU database). The sequences used for generating the reference OTUs in OTUXv2 have been derived using different pairs of primers used for 16S rRNA amplicon sequencing. As a result, the reference OTUs in the OTUXv2 database corresponds to the exact same sequence regions that would be picked up during an amplicon sequencing experiment. Consequently, the query reads may be used directly with OTUXv2 unlike OTUX approach where a preliminary pruning of reads was required to maintain parity between the length and type of the query and database sequences.
The reason to prefer "amplicon based" OTUXv2 approach over "V-region based" OTUX approach includes:
- Coherence between query and database sub-sequence sizes and locations.
- Retention of maximum possible sequence information generated through the experiment.
Please select the amplicon specific region of your choice from the dropdown below and then download the database.
The downloaded archived folder should have following files:
- FASTA file of containing sequences consituting the selected primers (amplicon specific sequences) corresponding to OTUX OTU IDs
- Taxonomy file, which provides sequence wise taxonomic information along with the OTUX IDs
- A mapping-back matrix for selected amplicon-specific sequence database (required for mapping back the OTUX IDs obtained in the abundance profiles to Greengenes IDs. Please refer to this page for detailed information about mapping-back matrices.
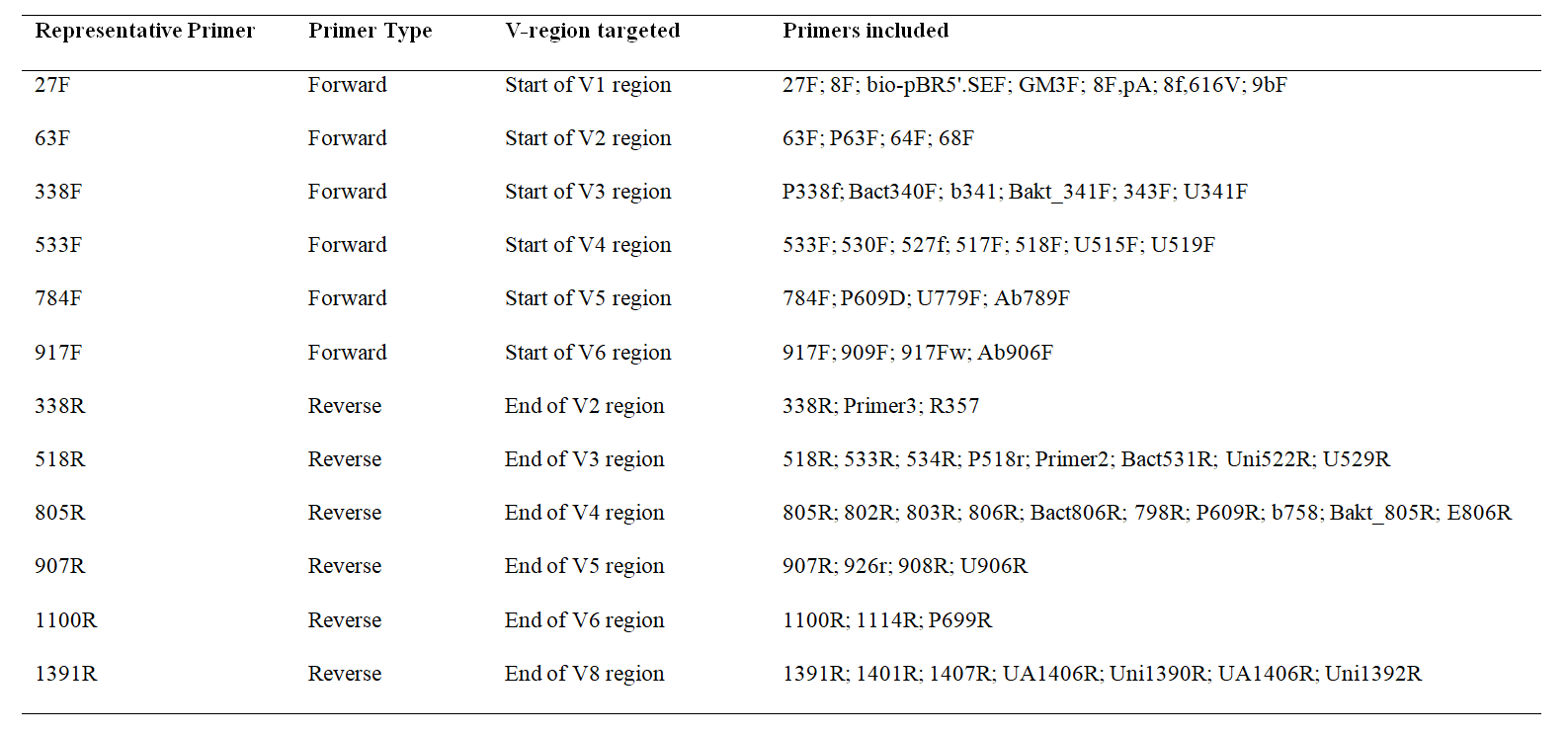
The following table provides more information regarding the primers used in OTUXv2 database.
The OTUXv2 databases have to be used using the exact procedure as OTUX database. Please check the help page for more details.
