iVikodak is an improved version of Vikodak (Nagpal et al., 2016) (PMID:26848568 ), developed with an objective of creating a one stop solution for predicting, analysing, comparing, and visualizing the Functional Potential of Microbial Communities.
iVikodak facilitates end-users to concomitantly infer, statistically analyse, and compare multiple microbial communities, and in the process generate a plethora of intuitive self-explanatory visual outputs in an automated fashion.
Global Mapper: The function inference module
Feature Analyzer: The comparative analysis module
Local Mapper: The pathway probing tool
Besides the three core modules there is also one auxiliary module as well:
A set of microbes in an environment can either exist as a consortium (i.e. with co-metabolic associations) or they might thrive independently. Considering these two possibilities (of bacterial co-existence), Global Mapper module of iVikodak provides to end-users the choice of two distinct algorithmic work-flows viz. 'Co-metabolism' and 'Independent Contributions'. The principle behind these two work-flows is as follows.
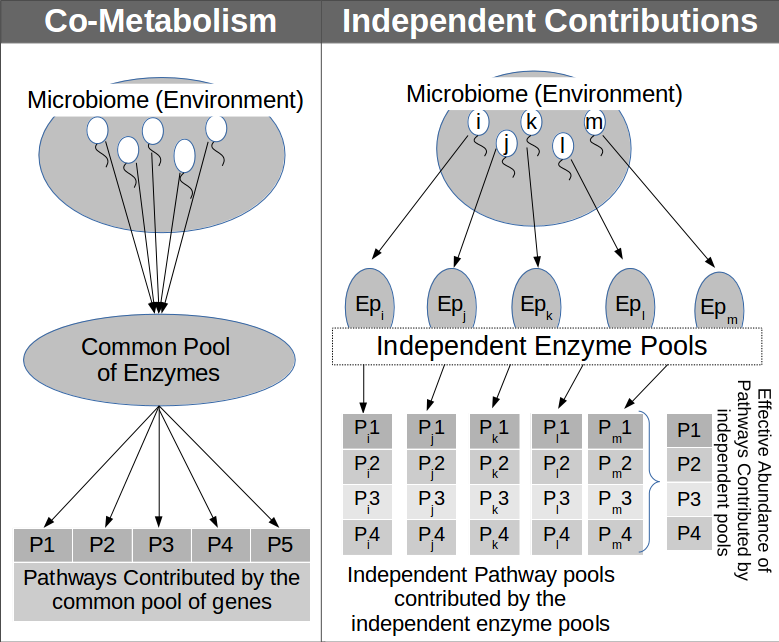
Co-Metabolism: The co-metabolism algorithm is based on the underlying assumption that the genes/ enzymes expressed by various microbes residing in an environment may pool together and contribute to the functioning of specific metabolic pathway(s). Therefore, the effective abundance of a metabolic pathway (in an environment with co-metabolising microbes) is expected to be a function of the total enzyme pool contributed by the co-metabolising microbes.
Independent Contributions: In contrast, the 'independent contributions' algorithm assumes the independent existence of microbes in the environment. Under this assumption, the effective abundance of a metabolic pathway is the sum total of the pathway abundances computed from individual microbes residing in that environment i.e the individual functional capacities of microbes independently contribute towards the effective presence or absence of a function
| Features | iVikodak | Vikodak | PICRUSt | Tax4Fun | |
|---|---|---|---|---|---|
| 1 | Multivariate inputs | ||||
| 2 | Default RDP compatibility | ||||
| 3 | Default GG compatibility | ||||
| 4 | Default Silva compatibility | ||||
| 5 | KEGG, pfam, cog, TIGRfam | ||||
| 6 | Single Pathway probing | ||||
| 7 | Gene Quorum Assumption | ||||
| 8 | Metadata acceptance | ||||
| 9 | Comparative Tests | ||||
| 10 | More than 2 environments | ||||
| 11 | Graphical Visualizations | ||||
| 12 | Function driven networks | ||||
| 13 | Personal Dashboards | ||||
| 14 | Portable Dashboards |